OVERVIEW OF LIST_MAPS RESEARCH PROGRAMME
The mechanisms by which Listeria monocytogenes adapts to a diverse range of environments remain largely unexplored but there is no doubt that the success of Listeria monocytogenes as a human foodborne pathogen relies on its overall ability to develop unique and sophisticated adaptation strategies which raises food safety issues.
The overall core of the List_MAPS research programme is to develop a systems biology approach to undersand how the conditions of the environment affect the capacity of Listeria monocytogenes to generate infection.
Four complex experimental sets-ups are used:
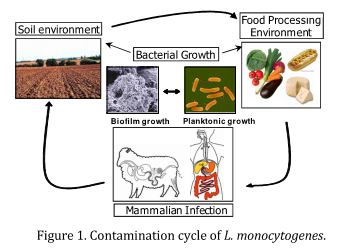
- soil (to follow listerial colonisation during growth of the plants)
- food-processing biofilms;
- food and food processing stress response (assess the effect of food composition on the response of Listeria monocytogenes)
- and humanised murine model
Different points are addressed:
- Deciphering transcriptomes and proteomes in complex environments;
- exploring adaptative mechanisms by reverse genetics;
- investigating interconnections between stress response, communication and RNome;
- Understanding the response to resident microorgarnisms;
- understanding how food matrix composition and environmental cues may affect the capacity of the pathogen to generate infection;
- Developing a transcriptome-based rapid tool, surrogate to animal models, to investigate the capacity of large collections of isolates to generate infection, in relation to environmental conditions: linking diversity and virulence;
- A commercial device will be adapted to develop a high throughput biofilm assay;
- Assessing light as disinfection treatment of foodstuff
IMPLEMENTATION
List_MAPS research programme is implemented in four Work packages that combine global analyses (transcriptome and proteome), classical molecular biology and physiological studies in controlled environments:
WP1: Data collection and integration in the specific environments
Objectives: Production of RNA and protein extraction in the various environments (task 1) database construction including TSS identification, RNA-seq and MACE analyses (task 2) and integration of transcriptomic and proteomic data (task 3).
Lead participant: GenXPro
WP2: Linking environmental cues and expression of virulence
Objectives: Virulence assays in relation to food constituents and chitin (task 1), exploitation of transcriptome data to analyse virulon expression in relation to environmental conditions (task 2), virulence assays of delection mutants (task 3).
Lead participant: University College Cork
WP3: Tools for evaluation of intra-specific phenotypic diversity
Objectives: Development and validation of an in silico assay surrogate to animal testing (task 1), development of a high throughput biofilm assay (task 2), analysis of the strain collection (task 3).
Lead participant: BioFilm Control
WP4: Systems biology approach
Objectives: Model construction (task 1), identification of target genes and reverse genetics approach (task 2), back and forth optimisation and dialog between the model and the bench (task 3).
Lead participant: INRA
INDIVIDUAL RESEARCH PROJECTS
The individual projects of the ESRs contribute to the overall research programme. Each sub-project contribute to the generation of data required for the regulatory network reconstruction and each individual project is designed to focus on one specific habitat and/or regulatory component. Through cooperation, individual contributions is magnified and as a consequence each individual project deepens its findings.
![]() This project has received funding from the European Union’s Horizon 2020 research and innovation programme under the Marie Sklodowska Curie grant agreement n° 641984
This project has received funding from the European Union’s Horizon 2020 research and innovation programme under the Marie Sklodowska Curie grant agreement n° 641984