Mouse models

We use mouse genetic approaches coupled with histology, brain anatomy, molecular and cellular biology, electrophysiology, animal behavior and bio-informatic approaches to understand principles of brain development and pathophysiology of brain malformation disorders.
Histology and brain anatomy
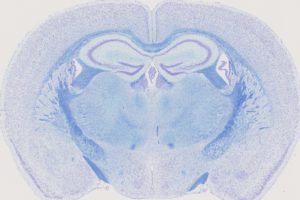
Coronal brain section of a mouse brain double-stained with Nissl and Luxol.
Detecting and examining the structural changes in diseased brains is a complex process because this tissue is so highly organized and requires extremely standardized histological methods for quantification of brain nuclei sizes. These methods play a key role in the investigation of cognitive disorders in animal models because, in many instances, brain anatomy can be considered an endophenotype of developmental brain disorders. Our group uses the digital slide scanner Hamamatsu Nanozoomer 2.0HT C9600 series producing whole 2D brain images at cell-level resolution and develops novel histological and imaging tools to understand brain structure and function in a three-dimension setting.
High-resolution episcopic microscopy
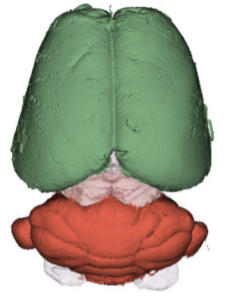
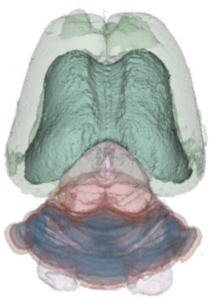
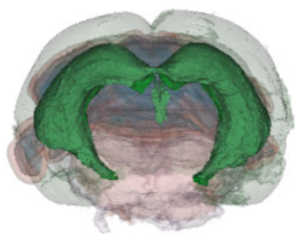
We have acquired high-resolution episcopic microscopy (HREM) technology for the study of whole embryonic and adult brains. HREM is a block-facing serial sectioning imaging technique in which samples are embedded in resin and sectioned within the body of a fluorescence microscope using an automated microtome. Therefore, HREM does not require the need of clearing or optical sectioning. The result is a multi-fluorescence 3D brain where brain regions are modelized. 24 brain structures are routinely segmented by manual annotations but we are in the process of developing artificial intelligence tools to automate segmentation (collaboration with Dr. Alain Lalande).
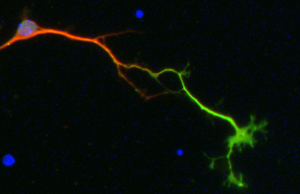
Cellular models
We use dissociated primary neuronal cultures for an easy access to living neurons in a simpler system for experimental manipulations and observations.
High-throughput electrophysiology
We use the multielectrode array technology to analysis neural networks and connectivity at unprecedented throughput. This system is ideal for phenotyping neuronal models in human and mouse and for drug screening applications.
Animal behavior and cognition
To address the functional consequences of genes whose disruptions yield NeuroAnatomical Phenotypes (NAP genes) including corpus callosum agenesis, on animal behavior and cognition, we typically assess motricity, locomotor activity, explanatory behavior, dexterity, forelimb muscular strength, hindlimb coordination, short- and long-term memory, spatial memory, innate tendency of mice to explore novel objects over familiar ones, sociability, anxiety- and depression-like behaviors. This is achieved using a variety of approaches: open-field test to study basic locomotor activity, hyperactivity and exploratory behavior; mouse reaching and grasping test to study forelimb reaching and grasping, as well as laterality in forelimb movement of the mice; grip strength test to evaluate forelimb muscular strength; notch bar test for hind limb coordination; Y-maze test to evaluate short-term working memory, based on the innate curiosity of the mice to explore an arm that has not been previously explored; novel object recognition test for the innate tendency of mice to explore novel objects over familiar ones; morris water maze to evaluate spatial memory; crawley recognition test to assess social recognition and interaction; hot plate test to evaluate thermal pain response; elevated plus maze test to study anxiety-like traits; forced swim and tail suspension tests to study depression-like behaviors.
Bioinformatics
We use bioinformatics to test whether neuroanatomical phenotypes co-occur within related brain regions by analyzing correlations. To group genes based on their phenotypic similarities, we use clustering methods such as t-Distributed Stochastic Neighbor Embedding (t-SNE) as well as a hierarchical method of brain parameters relations. We use genomic data sources including gene expression, protein-protein interactions (PPIs) and functional mouse and human datasets to identify functional convergence amongst genes associated with neuroanatomical defects. By this means, we can discover new candidate pathways for therapeutic intervention. To assess disease relevance, we compare evolutionary properties and sequence changes in the NAP genes. Several measures are considered including the probability of being loss of function intolerant (pLI) and the rate of synonymous (dS) and non-synonymous (dN) gene substitutions.
